The following article contains excerpts from a research paper submitted as part of a term project in Natural Language Processing. The main purpose of this project was to analyze and implement a previously published research paper of our choosing relating to modern solutions for NLP named entity recognition. I chose to implement and analyze GRAM-CNN originally implemented by the University of Florida.
The Problems of Scientific NER
A common task in NLP, known as Named Entity Recognition, consists of identifying key words or concepts in a text as part of a classifiable entity. Generally this problem is tackled through statistical modeling, feature dictionaries, hand-crafted rules, etc. However, when applied to a complex textual domain like dense biological texts these models under-perform. While they may have some success with standard texts, these models are: expensive to produce, highly specialized to their original domain, and not generizable to new domains, new vocabulary, or non-standard usage of old concepts. Meanwhile, biology based text is made up of multiple languages like Latin and Greek, named entities that compose other named entities, long words and phrases used with varying frequency, and a constantly increasing vocabulary of these words and phrases. The most commonly used methods for Bio NER involve machine learning techniques applied along with the statistical modeling method known as conditional random fields (CRF). However, while CRFs are ideal for modeling, and subsequently labeling, sequences using the intermixed neighboring context of words, they heavily rely on features of any given input. These may include pre-defined “orthographic,morphological, linguistic-based, conjunctions and dictionary-based” rules that define the context and usage of words in the text. Like the previously mentioned methods, these features are generally hand-crafted through expensive human analysis and not transferable to new text. However, we can attempt to learn these features using Deep Learning techniques, combined with the proven use of CRF, and an appropriate dataset.
Original GRAM-CNN Paper
The original authors of GRAM-CNN theorized that through the use of deep convolutional layers, pre-trained embeddings, and syntactical features they could form an accurate and generizable model applicable to multiple forms of biological text . Their main goal was to provide a representation model that could fill in the gaps when a new or misspelled word appears in the text. To achieve this the authors needed an initial representation model for each word. Rather than use a sparse / orthogonal representation like one-hot encodings that capture none of the meaning or context behind words, the authors chose Word2Vec.
Word2Vec Representations
Word2Vec is a probability based method that learns weights that can predict a probability distribution for each context word in a fixed-size sliding window given a center word as a conditional value. This method iterates over the full text and updates a series of weights for the center ![]() and context words
and context words ![]() based on their accuracy to predict the center
based on their accuracy to predict the center ![]() and context word
and context word ![]() over all words in the vocabulary. The overall goal is to maximize the total likelihood, which we can define as the chained conditional probability of all the context words in a window, chained over the entire text, using our learned representations. Unlike the standard one-hot encoding method, this method allows for a learned vector representation of the word that is lower in dimension, encompasses more than just the discrete label for the word, and is transferable to other tasks. However, this method is limited by the vocabulary found in the training corpus, and is unable to generalize towards completely unseen or mispelled words. To address this the authors use a character level model built up using multi-layer convolutions.
over all words in the vocabulary. The overall goal is to maximize the total likelihood, which we can define as the chained conditional probability of all the context words in a window, chained over the entire text, using our learned representations. Unlike the standard one-hot encoding method, this method allows for a learned vector representation of the word that is lower in dimension, encompasses more than just the discrete label for the word, and is transferable to other tasks. However, this method is limited by the vocabulary found in the training corpus, and is unable to generalize towards completely unseen or mispelled words. To address this the authors use a character level model built up using multi-layer convolutions.
![]()
![Rendered by QuickLaTeX.com \[ L(\theta) = \prod_{t=1}^T \prod_{W_o \epsilon W(t)} P(W_o|W_t; \theta) \]](https://www.pseudobrilliant.com/wp-content/ql-cache/quicklatex.com-422a2e18fd009b4ca808e987e91635e3_l3.png)
GRAM-CNN Approach
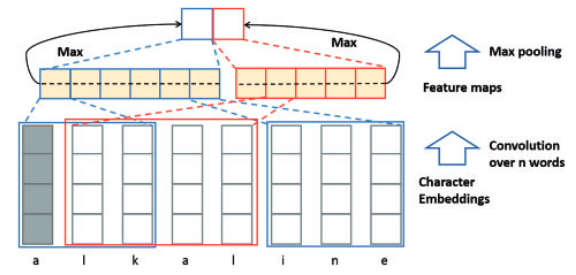
Essentially, the authors attempt to build a word representation by building up from simple character groupings, of multiple sizes, to higher order combinations that may represent the word. The authors posit that combining this representation with the word embedding may fill in the gap when a word is missing or misspelled. The authors implement this concept using stacked 2D convolutional layers of different kernel and feature sizes. By applying these convolutions to an embedding representation of the input, they can achieve a mixture of the word representation at different kernel sizes / character distance. They use max pooling layers after each convolution in order to extract only the most impactful features. Once the convolutions are complete, they concatenate the resulting feature maps into a sequence the same size as the original input. Over this sequence they also concatenate the Word2vec representation for each word and a index representation of the syntactical role it plays (i.e part of speech). Once each set of features is concatenated into a total representation the authors employ another 2D convolutional stack, similar to the first, in order to simulate N-gram calculations. In a standard implementation, the main goal is to model the language used in the training text by grouping words at varying sizes 1 to N-1, and predict the next word for the group at size N. The authors use the total embedding as input to a convolutional stack of varying kernel sizes in order to mimic a form of this concept. Max pooling is again used to reduce the resulting feature map for each time step to only the most useful features.
CRF Sequence Predictions
The authors then employ a linear-chain conditional random field to generate the most probable sequence of labels. The author’s implementation of CRF uses a unitary potential for each entity label and a binary potential for transition between label types. The(T) matrix provides transition score for moving from the previous tag to the current, while the P matrix provides the score of the current word (i) matching to a label (y). This forms the objective function which we train for by maximizing the log likelihood probabilities of labels of sequences produced by a softmax function. The CRF layer allows the model to make label predictions by taking into account the label score of a word and the words around it.
![]()
GRAM-CNN Self Implementation
My own implementation followed the original description of the model as described in the paper. The code implementation provided by the authors did not entirely match what was described, and contained multiple complex components not mentioned in the original paper (highway, batch normalization, etc). Instead I focused my efforts on following the spirit of the paper and attempting to replicate the results they achieved on the NCBI disease corpus dataset, with a few modifications of my own. As part of this project, I re-implemented the generation of the tokenized and labeled NCBI dataset. Which led to the discovery that the original authors had not in fact labeled their dataset properly, and that the NCBI dataset they had trained on only consisted of 2 named entities; A fact which may account for the high ~85\% accuracy their model achieved. Instead, I took the 9 seperate disease classifcations available in the dataset as my target NER labels. The dataset was tokenized according to the IOBS standard and padded to a max sequence length size. For each word, If the word existed in vocabulary of the pre-trained PubMD word2vec embedding the authors provided then the index for that word was made part of the sequence. Otherwise, an ‘unseen’ character was placed at the questionable spot in the sequence. The same sequence of max length would be generated with an index for each character present, padded to a common max character length as well. The word2vec index sequence, character index sequence, and a POS sequence generated by nltk toolkit would then become the input for the model.
The model creation was done with Keras instead of the original tensorflow in order to provide a simplified implementation. As shown in the figure above this implementation is made up of two main convolutional blocks joined together by concatenations of their resulting features. The first block accepts the character level input, performs the character n-gram comvolutions and produces a concatenated feature vector over the full max sequence. This sequence is concatenated with word2vec representation for each word in the sequence. At both concatenation levels a masking sequence is applied so that the model does not learn from unseen sequence characters or words. The word and character feature concatenations are then fed to the n-gram convolutional block. Again the resulting features are concatenated over each step, along with the POS input values at each step, before finally being provided to the CRF for sequence classification. The model was trained with a CRF loss objective and a Adam optimizer for dynamic learning rates during training. The addition of masking layers, dropout layers, and Adam optimization are my contributions designed to address the missing highway layer and batch normalization while still maintaining the original structure.
Results and Conclusion
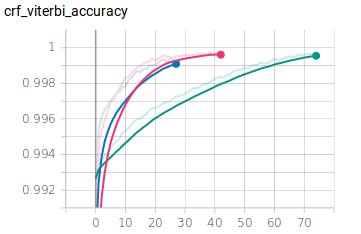
My implementation was trained over 75 epochs spanning 80\% the NCBI training set for training and 20\% for training validation. I set the model to train for a longer range of epochs, but used early stopping relative to the validation set to limit training from over fitting on the training set. The crf accuracy achieved excellent results in both training and validation sets during training, but the resulting accuracy for multi-NER sequence predictions on the test set was only 64\%. I applied this same architecture to the same dataset, but with one NER labels like in the original paper, and achieved a higher accuracy result of 70\%. These scores are still lower than the results achieved by the original paper. This discrepancy could possibly be explained by the original paper’s use of more feature layers with varying kernel sizes, and the application of highway layers to achieve better training for these deeper networks. The multi-NER results also show that the CompositeMention and SpecificDisease labels performed particularly poorly in comparison with the rest of the labels. These types of labels are more compositional than the rest, meaning that the words involved may be incorrectly classified as part of the DiseaseClass. Adding feature layers at different kernel sizes of a larger size may allow the model to learn more complex combinations of words, possibly extracting more combination features, leading to increased accuracy. However, the current model uses up most of my NVIDIA GTX 1070s 8GB of RAM, meaning that deeper training would require more resources than I have available. In any case, these concepts may give further credence to the theory that the original paper’s accuracy may be best attributed to it’s depth and deep training techniques. These results still seem to imply that the general representation mix and match concepts of the original paper are applicable and can result in accurate NER labeling.




Leave a Comment